Microbiomics has rapidly advanced in recent years, yielding numerous new discoveries. However, the field lacks a comprehensive assessment of the reliability of its methods and protocols. Many researchers have highlighted issues with data reproducibility across labs and the challenges of managing the considerable variability inherent in the complex microbiomics workflow.
NGS microbiomics workflows consist of multiple intricate steps, often complicated by biases and errors occurring throughout. Figure 1 outlines the typical challenges encountered at each stage of the microbiomics workflow. Even freezing, which is considered the gold standard, can potentially suffer from bias caused by freeze-thaw cycles. Read our blog to learn how bias can arise in every step of your workflow.
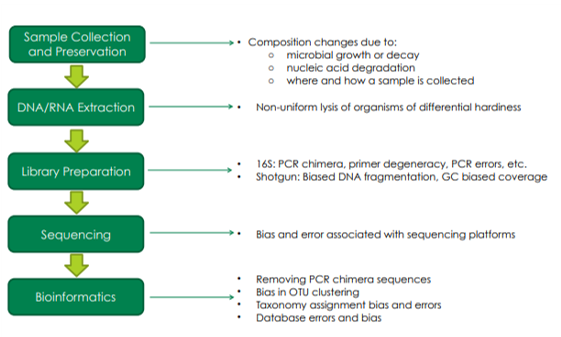
Figure 1. Potential sources of bias or error throughout the entire microbiomics workflow.
The ZymoBIOMICS® portfolio represents Zymo Research's response to the demand for improved reliability in microbiome analysis. It comprises various products designed to tackle key challenges in the microbiomics workflow, such as microbial community standards, sample collection devices, DNA/RNA isolation kits, and library preparation kits. Additionally, Zymo provide a comprehensive microbiomics sequencing service for customers who opt to delegate technical processing.
The microbial community standards from Zymo Research allow scientists to validate and standardise their methods and have confidence in their results. Zymo certify all of their microbial standards to have <0.01% microbial contamination (by DNA abundance). Read our blog about how Zymo's microbial community standards have become the most cited in the industry.
Microbial community standards available
| ZymoBIOIMCS standards | Microbial Community Standard II | Spike-in Controls | Fecal Reference | |||||
|---|---|---|---|---|---|---|---|---|
| Formats | Microbial cells | Isolated DNA | Microbial cells | Isolated DNA | High microbial load | Low microbial load | Natural material, whole cell | |
| Detects bias in | Complete workflow | Library prep and sequencing | Complete workflow | Library prep and sequencing | Individual samples | Individual samples | Complete workflow | |
| Recommended use | General benchmarking and microbiome profiling control control | Assess detection limit and sensitivity | Absolute quantification of high bacterial load samples e.g. faeces | Absolute quantification of low bacterial load samples e.g. sputum | Method comparison and process control | |||
For long read library prep and sequencing, a high molecular weight DNA standard is available. This isolated DNA standard can be used as a positive control for general benchmarking and microbiome profiling.
Microbiomics & metagenomics portfolio
Cambridge Bioscience offers a comprehensive solution for microbiomic and metagenomic research from collection to analyses. See the full range.




