The ZymoBIOMICS™ Spike-in Controls from Zymo Research are in situ positive, whole cell exogenous controls that enable cell number measurements from high or low bacterial load samples using next-generation sequencing (NGS). Comprised of bacterial species alien to the human microbiome, these standards are ideal for ensuring each sample is accurately quantified.
The ZymoBIOMICS™ Spike-in Control I (High Microbial Load) consists of accurately quantified cell numbers of two bacterial strains, Imtechella halotolerans (gram-negative) and Allobacillus halotolerans (gram-positive). Representing different cell recalcitrance, these strains can expose potential bias during DNA extraction and enable absolute cell number quantification in microbiome measurements.
The ZymoBIOMICS™ Spike-in Control II (Low Microbial Load) consists of three bacterial strains, Truepera radiovictrix, Imtechella halotolerans and Allobacillus halotolerans. Featuring gram-negative, gram-positive and high GC content, lysozyme resistant bacterial strains, this standard represents different challenges in NGS-based microbiome analysis. With accurately quantified cell numbers and a log abundance distribution, this standard enables absolute cell number quantification in cases such as pathogen load detection.
ZymoBIOMICS Spike-in Control I workflow

- Add ZymoBIOMICS™ Spike-in Control I to sample
- DNA extraction, library preparation & Next-Generation Sequencing
- Bioinformatic analysis to measure absolute microbial abundance
Defined composition
|
Spike-In Species
|
Per Prep (20 µl)
|
||
|---|---|---|---|
|
Cells
|
16S Copies
|
Total DNA (ng)
|
|
|
Imtechella halotolerans
|
2 x 107
|
6.0 x 107
|
67.2
|
|
Allobacillus halotolerans
|
2 x 107
|
1.4 x 108
|
58.2
|
Quantify absolute abundance of microbes
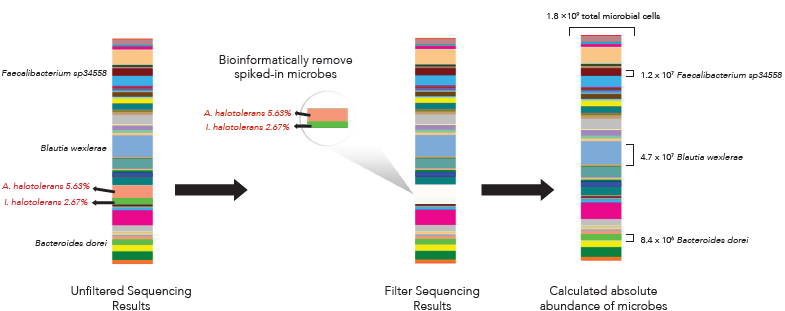
The ZymoBIOMICS Spike-in Control I was added to a fecal sample. Total DNA was then extracted using the ZymoBIOMICS DNA Miniprep Kit, a library was prepared and then analyzed by 16S targeted Next-Gen Sequencing. From the resulting data, the percentage of Imtechella halotolerans and Allobacillus halotolerans in the total sample composition was then calculated. The resulting percentage was paired to the defined Spike-in I cell input and used to calculate the absolute abundance for organisms in the sample based their sample composition percentages.
Material available for download
ZymoBIOMICS™ Spike-in Control I (High Microbial Load) protocol
ZymoBIOMICS™ Spike-in Control II (Low Microbial Load) protocol
Products
Note: product availability depends on country - see product detail page.
| Details | Cat number & supplier | Size | Price |
| ZymoBIOMICS Spike-in Control II (Low Microbial Load) (500 µl x 1) D6321 · Zymo Research | D6321 Zymo Research |
25 preps |
£160.00
25 preps
view
|
| ZymoBIOMICS Spike-in Control II (Low Microbial Load) (500 µl x 10) D6321-10 · Zymo Research | D6321-10 Zymo Research |
250 preps |
£812.00
250 preps
view
|
| ZymoBIOMICS® Spike-in Control I (High Microbial Load) (500 µl x 1) D6320 · Zymo Research | D6320 Zymo Research |
25 preps |
£160.00
25 preps
view
|
| ZymoBIOMICS® Spike-in Control I (High Microbial Load) (500 µl x 10) D6320-10 · Zymo Research | D6320-10 Zymo Research |
250 preps |
£812.00
250 preps
view
|