The ZymoBIOMICS™ Microbial Community Standard II is a well-defined, mock microbial community ideal for assessing the detection limit of microbiomics workflows and as a positive control for routine sequencing. Consisting of eight bacterial and two fungal strains mixed to create a log-distributed abundance, this standard is accurately characterised and contains negligible impurities. 75 µl of the standard contains ~100 cells of Staphylococcus aureus, the organism of lowest abundance. If needed, the standard can be spiked into a sample matrix (e.g. soil and blood) to mimic real samples of interest.
Benefits of using this log distribution standard
- Well defined & characterised
- Less than 0.01% impurities
- Ideal for QC of microbiome measurements
- Assess the accuracy of microbiome measurements
Also available is the ZymoBIOMICS™ Microbial Community DNA Standard II, which consists of a mixture of genomic DNA of eight bacterial and two fungal strains and is ideal for assessing the performance of microbiomics workflows or as a positive control for routine QC. The microbiobial standard is accurately characterised and contains negligible impurities (<0.01%).
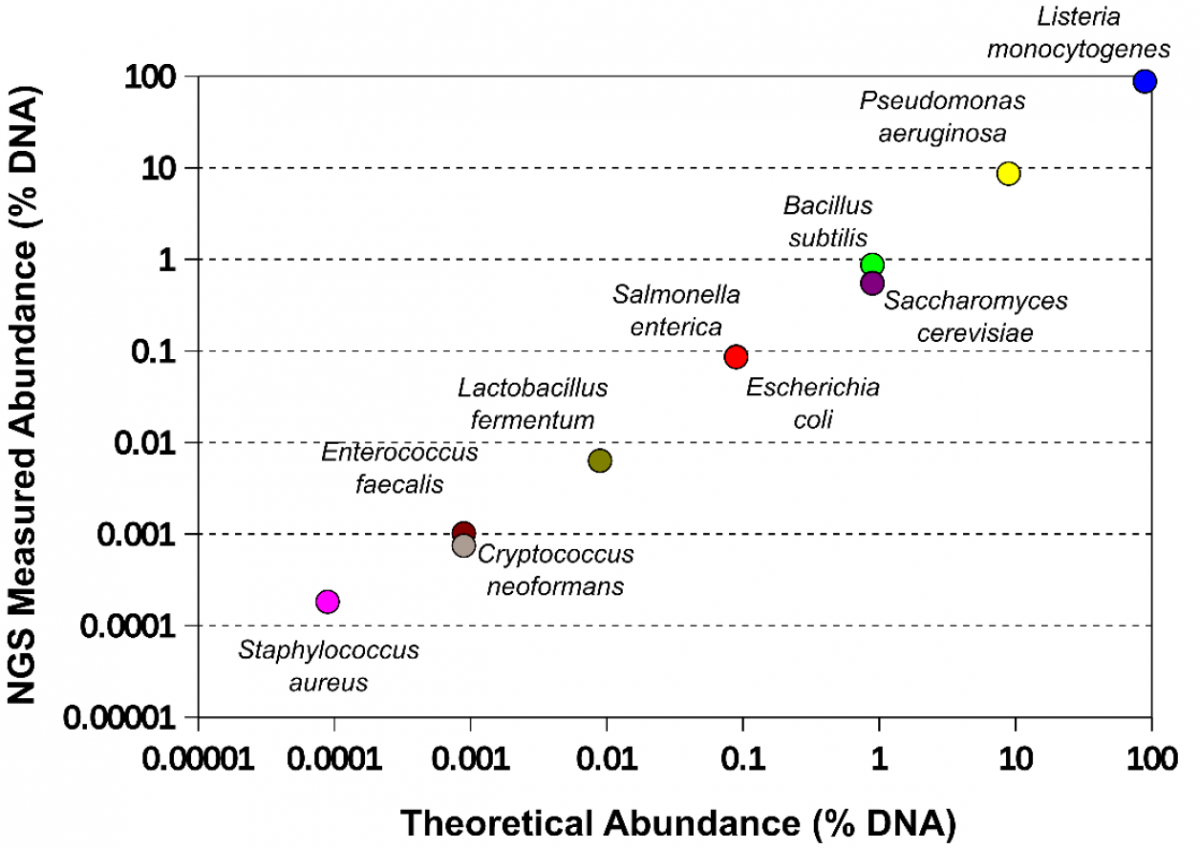
The microbial composition of the standard measured by NGS shotgun sequencing as compared to the defined composition. After mixing, the microbial composition of the standard was confirmed using deep Illumina® shotgun sequencing. Briefly, the genomic DNA was extracted using the ZymoBIOMICS™ DNA Miniprep. Library preparation was performed using an in-house protocol. Shotgun sequencing was performed using Illumina HiSeq™ or MiSeq™. Microbial abundance was estimated based on the number of reads that were mapped to references genomes of the organisms.
Microbial composition
| Species | Defined composition (%) | ||||
|---|---|---|---|---|---|
| Genomic DNA | 16S only1 | 16S & 18S1 | Genome copy2 | Cell number2 | |
| Listeria monocytogenes | 89.1 | 95.9 | 91.9 | 94.8 | 94.9 |
| Pseudomonas aeruginosa | 8.9 | 2.8 | 2.7 | 4.2 | 4.2 |
| Bacillus subtilis | 0.89 | 1.2 | 1.1 | 0.7 | 0.7 |
| Saccharomyces cerevisiae | 0.89 | NA | 4.1 | 0.23 | 0.12 |
| Escherichia coli | 0.089 | 0.069 | 0.066 | 0.058 | 0.058 |
| Salmonella enterica | 0.0089 | 0.07 | 0.067 | 0.059 | 0.059 |
| Lactobacilus fermentum | 0.0089 | 0.012 | 0.012 | 0.015 | 0.015 |
| Enterococcus faecalis | 0.00089 | 0.00067 | 0.00064 | 0.001 | 0.001 |
| Cryptococcus neoformans | 0.00089 | NA | 0.0014 | 0.00015 | 0.00007 |
| Staphylococcus aureus | 0.000089 | 0.0001 | 0.0001 | 0.0001 | 0.0001 |
1 The theoretical composition in terms of 16S (or 16S and 18S) rRNA gene abundance was calculated from theoretical genomic DNA composition with the following formula: 16S/18S copy number = total genomic DNA (g) × unit conversion constant (bp/g) / genome size (bp) × 16S/18S copy number per genome. Use this as reference when performing 16S targeted sequencing.
2 The theoretical composition in terms of genome copy number was calculated from theoretical genomic DNA composition with the following formula: genome copy number = total genomic DNA (g) × unit conversion constant (bp/g) / genome size (bp). Use this as reference when inferring microbial abundance from shotgun sequencing data based on read depth.
3 The theoretical composition in terms of cell number was calculated from theoretical genomic DNA composition with the following formula: cell number = total genomic DNA (g) × unit conversion constant (bp/g) / genome size (bp)/ploidy.
Microbiomics & metagenomics portfolio
Cambridge Bioscience offers a comprehensive solution for microbiomic and metagenomic research from collection to analyses. See the full range.
Material available for download
ZymoBIOMICS™ Microbial Community Standard II (Log Distribution) protocol
ZymoBIOMICS™ Microbial Community Standard II (Log Distribution) SDS
ZymoBIOMICS™Microbial Community DNA Standard II (Log Distribution) protocol
ZymoBIOMICS™ Microbial Community DNA Standard II (Log Distribution) SDS
Products
Note: product availability depends on country - see product detail page.
| Details | Cat number & supplier | Size | Price |
| ZymoBIOMICS Microbial Community Standard II..(Staggered, Cellular Mix), 750 µl D6310 · Zymo Research | D6310 Zymo Research |
10 preps |
£489.00
10 preps
view
|
| ZymoBIOMICS Microbial Community DNA Standard II (Log Distribution) 220 ng, 20µl D6311 · Zymo Research | D6311 Zymo Research |
220 ng |
£251.00
220 ng
view
|