FFPE samples play a crucial role in cancer research and oncology, offering a stable sample type essential for experimental studies, diagnostics, biomarker identification, and drug development target studies. These collections hold immense potential for advancing the understanding and treatment of various human diseases. FFPE samples are highly valuable yet contain compromised, low-quality, and degraded RNA, making them challenging to work with from the point of view of next-generation sequencing.
To address these challenges, specialised RNA-Seq library preparation kits are required. These kits must possess exceptional sensitivity and the capability to generate libraries from degraded RNA samples. For differential expression analysis of FFPE samples, both whole transcriptome sequencing (WTS) and 3′ mRNA-Seq are effective methods. Lexogen offers tailored solutions for both approaches, ensuring reliable and accurate results.
The SPLIT One-step FFPE RNA Extraction and Lexogen FFPE Library Prep Kits provide a seamless end-to-end workflow for efficiently handling the most challenging FFPE material. This comprehensive solution includes data analysis on Kangooroo, Lexogen’s user-friendly web-based platform for NGS data processing.
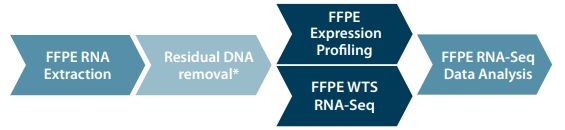
Figure 1. Lexogen's FFPE RNA library preparation workflow.
FFPE gene expression profiling
Lexogen’s QuantSeq FFPE 3’ mRNA-Seq gene expression kit offers a fast and cost-effective solution, specifically designed and validated for FFPE samples from a variety of tissues and organisms. Utilising 3’ mRNA-seq technology, it targets the polyA tail of polyadenylated transcripts, ensuring one read per transcript, significantly lowering sequencing depth requirements, as well as costs for data analysis and storage. Additionally, 3’ mRNA-Seq eliminates the need for pre-processing steps like mRNA enrichment or rRNA depletion, streamlining the workflow and reducing both processing steps and consumable usage.
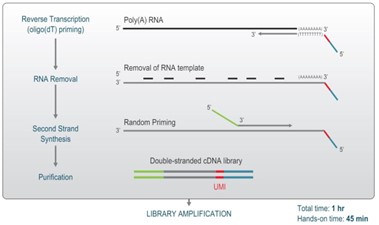
Figure 2. QuantSeq FFPE library preparation workflow.
FFPE whole transcriptome sequencing
Paired with RiboCop for enzyme-free rRNA depletion, CORALL FFPE Whole Transcriptome RNA-Seq enables in-depth, comprehensive analysis of the entire transcriptome, covering all RNA molecules, including non-polyadenylated and long non-coding RNAs. It also facilitates isoform detection, analysis of fusion transcripts, alternative splicing events, transcriptome assembly, and more. Whole transcriptome sequencing studies often follow gene expression analysis to unravel molecular mechanisms underlying the changes in expression.
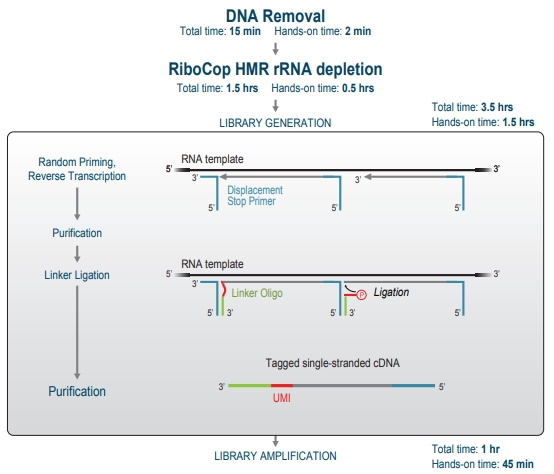
Figure 3. CORALL FFPE whole transcriptome library preparation workflow.
Choosing which analysis is best for your FFPE samples
This choice depends on the experimental aim, i.e., if quantitative or qualitative data is required, on the RNA molecules of interest (e.g., mRNA. lncRNA, etc.), and economical factors around the project. For example, budget, sample availability, and data storage can also be important aspects to consider when deciding for one approach over another. There is a high degree of overlap in gene detection for FFPE samples processed with 3′ mRNA-Seq (using QuantSeq FFPE) and fragmentation-free whole transcriptome RNA-Seq (using CORALL FFPE). The results demonstrate that 3′ mRNA-Seq and whole transcriptome RNA-Seq are equally effective and suitable for looking at gene expression from archived FFPE samples.
Products
Note: product availability depends on country - see product detail page.
| Details | Cat number & supplier | Size | Price |
| QuantSeq FFPE 3’ mRNA-Seq Library Prep Kit FWD with UDI 12 nt Set A1, (UDI12A_0001-0096) 222.96 · Lexogen | 222.96 Lexogen |
96 preparations |
£3249.00
96 preparations
view
|
| CORALL FFPE Whole Transcriptome RNA-Seq Library Prep with UDI 12 nt Set A1, (UDI12A_0001-0096) 219.96 · Lexogen | 219.96 Lexogen |
96 preparations |
£3784.00
96 preparations
view
|
| RiboCop (HMR) and CORALL FFPE Whole Transcriptome RNA-Seq Library Prep with UDI 12 nt Set A1, (UDI12A_0001-0096) 233.96 · Lexogen | 233.96 Lexogen |
96 preparations |
£8988.00
96 preparations
view
|

