- In this section:
- Contact our Zymo custom service specialists
Full-length amplicon sequencing provides unrivalled taxonomic species-level resolution by sequencing the full rRNA gene, not just the hypervariable regions within it. rRNA genes range in size from 1-2kb, sizes short-read platforms cannot capture, so the resulting amplicons are sequenced using the latest PacBio Revio platform. This method allows for the identification and resolution of closely related species that would not be resolved with other metagenomic sequencing methods. Zymo is a PacBio certified service provider.
What you can expect from Zymo’s services
- Quick turnaround time: results in as little as 3 weeks
- Unrivalled species resolution
- Consultation and one-to-one support
- Unrivalled data quality
- Stringent QC using microbial standards
- Reference database curated in-house
Comprehensive bioinformatics analysis report
The full-length 16S rRNA sequencing service report provides compositional bar plots, composition heatmaps, alpha-diversity and beta diversity plots, and statistical group comparison with LEfSe.
Continuous monitoring for standardisation and consistency
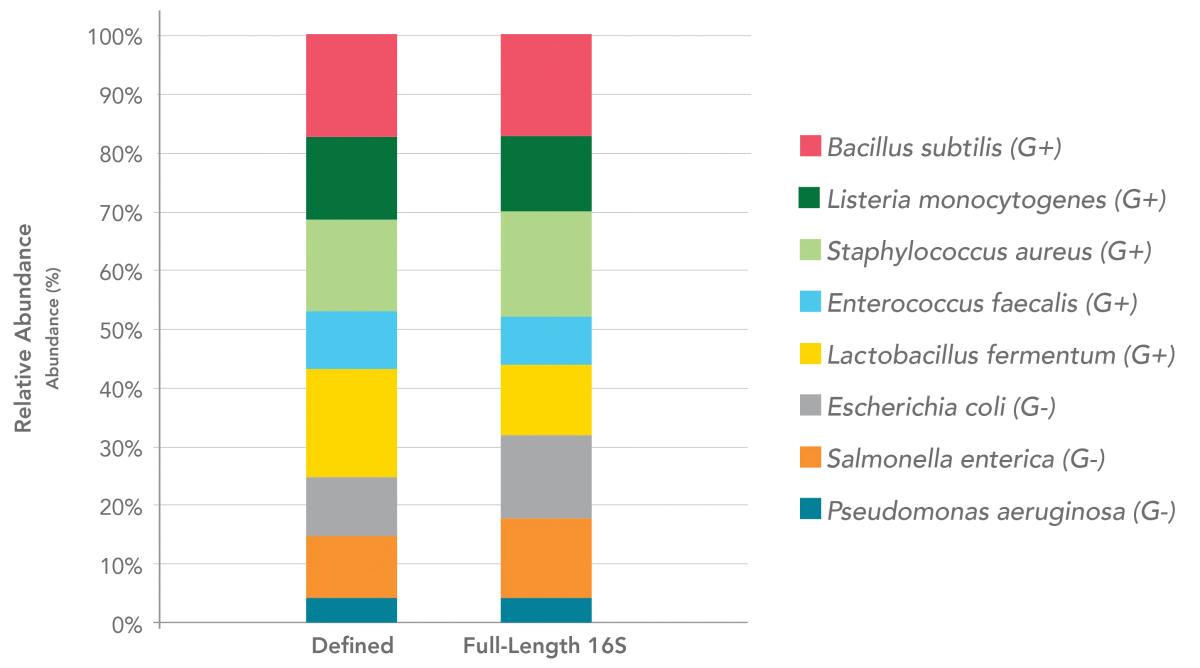
Zymo’s full-length 16S sequencing pipeline results in the same community profile as expected for their Microbial Community DNA Standard. The pipeline is consistently monitored with Zymo’s own community standards, which have been cited in over 1000 publications and have helped the microbiomics research community move towards standardisation and reproducibility.
Full-length amplicon sequencing technical specifications
| Sequencing platform | PacBio Sequel Ile |
|---|---|
| Sequencing kit | SMRT Cell 8M |
| Target amplicon | The full-length 16S rRNA gene is amplified with 27f (AGRGTTYGATYMTGGCTCAG) and 1492r (RGYTACCTTGTTACGACTT) |
| Accepted sample types | Extracted genomic DNA or raw samples |
| DNA extraction |
ZymoBIOMICS 96 Magbead DNA (D4308) ZymoBIOMICS DNA Miniprep (D4300) |
| Library prep | Zymo follows PacBio recommended protocol (https://www.pacb.com/wp-content/uploads/Procedure-checklist-Amplification-of-bacterial-full-length-16S-rRNA-gene-with-barcoded-primers.pdf) |
| Quality control | A positive control for DNA extraction (ZymoBIOMICS Microbial Community Standard, D6300) and a positive control for library preparation (ZymoBIOMICS Microbial Community DNA Standard, D6305) are included in each project as a quality control. Results are reported in the final project report. Negative extraction controls and negative qPCR controls are also monitored with real time PCR and can be analyzed upon request. |
| Sequencing analysis | Amplicon Sequence Variant (ASV) based analysis with DADA2 and Qiime 1.9 is used to provide higher accuracy and better resolution of data. |
| Reference database | As publicly available databases (e.g. Greengenes, UNITE) contain a multitude of errors, inconsistent annotations, and poor sequence quality, we utilized an in-house curated reference database. |
| Data visualisation | Taxa abundance barplot, taxa abundance heatmap, ASV abundance heatmap, alpha diversity boxplot and rarefaction curves, beta diversity 3D Emperor plot, Taxa2ASV decomposer |
| Statistical analysis | LEfSe analysis uncovers taxa/traits that are differentially distributed among defined sample groups. |
| Data storage | Raw sequencing data and analysis report are stored for up to 3 months |

