- In this section:
- Contact our Zymo Research specialists
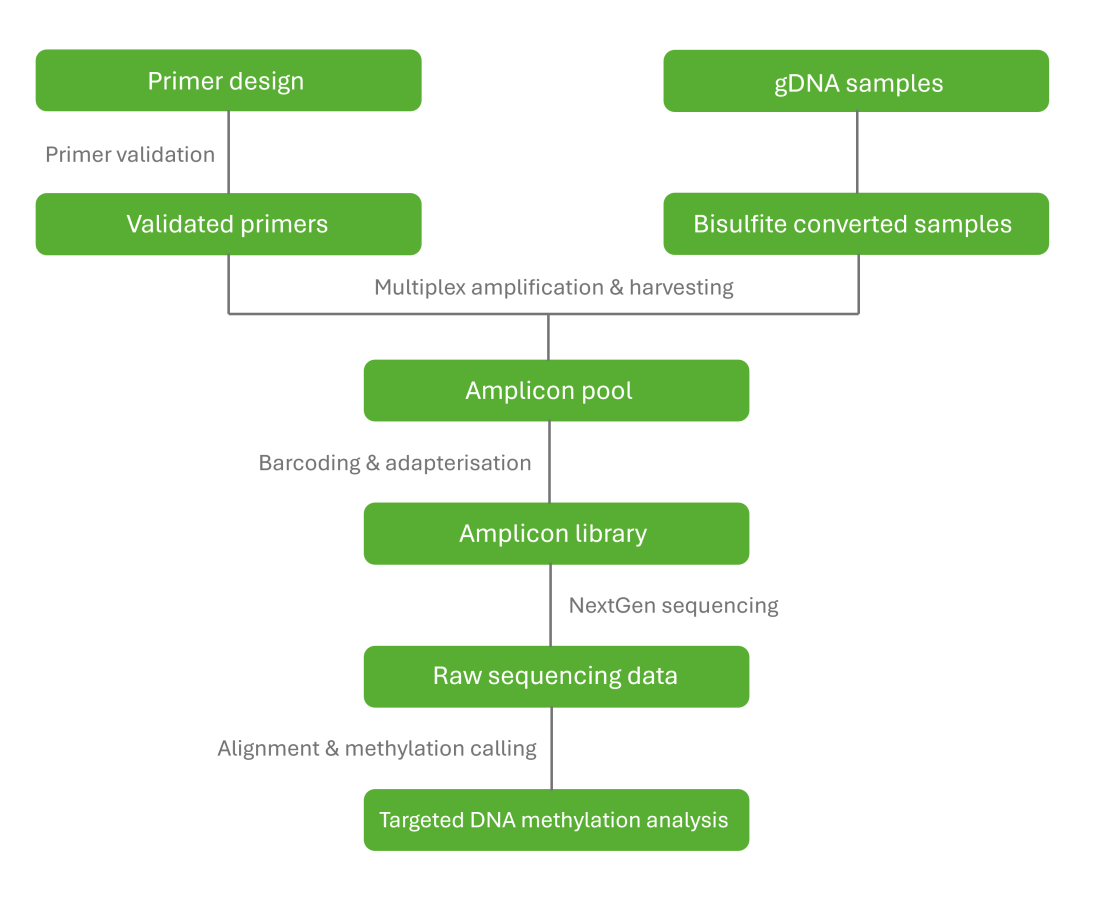
Targeted bisulfite sequencing offers targeted DNA methylation analysis of specific regions of interest within the genome. Methylation status of cytosines is determined by converting unmethylated cytosines to uracil using bisulfite treatment, while methylated cytosines remain unchanged. Using next-generation sequencing methods to sequence the treated DNA, methylated cytosines can be identified and methylation patterns analysed.
Zymo’s targeted bisulfite sequencing service is supported by their expert team and offers a personalised approach with primer design and validation for your region of interest. Send them your samples and regions of interest and they will take care of the rest.
What you can expect from Zymo’s targeted bisulfute sequencing service
- Tailored primer design and validation
- High throughput, targeted amplification
- Simplified data interpretation
- User-friendly analysis report
- Comprehensive DNA methylation analysis
Comprehensive bioinformatics analysis
Zymo’s targeted bisulfite sequencing service report provides quality histograms, bismark alignment rates, and CpG library coverage. Reports also provide full access to all raw sequencing data and Zymo’s team of bioinformaticians is on-hand to assist you with data interpretation and getting the most out of your results.
Targeted bisulfite sequencing workflow