- In this section:
- Contact our Zymo Research specialists
DNA methylation analysis involves bisulfite treatment of DNA to convert unmethylated cytosines to uracil, leaving methylated cytosines to be detected by next generation sequencing. This method detects epigenetic modifications such as 5-methylcytosine and 5-hydroxymethylcytosine, which play important roles in gene regulation, development, and genomic stability.
What you can expect from Zymo’s DNA methylation analysis services
- High-quality data ensured by spike-in controls
- A team of experts with 20 years of epigenetic experience
- Data analysis report with publication-ready figures
- Compatibility with low input
Zymo’s services begin with a free consultation to discuss your project and pipeline customization, are compatible with low input and any species with a reference genome, and finish with a user-friendly analysis report with publication-ready figures. Zymo offers two types of DNA methylation analysis:
Whole genome bisulfite sequencing
Whole genome bisulfite sequencing (WGBS) provides full coverage of the methylation status of the whole genome, including the less common contexts of CHG and CHH.
Reduced representation bisulfite sequencing
Reduced representation bisulfite sequencing (RRBS) is a DNA methylation analysis technique that focuses on methylation within CpG islands of the genome, the most common sites of methylation in mammalian species. This more targeted approach is considered a more cost-effective alternative to WGBS if full methylome analysis isn’t required.
| Basic report includes | Full report includes |
|---|---|
|
|
High-quality guarantee with spike-in controls in every sample
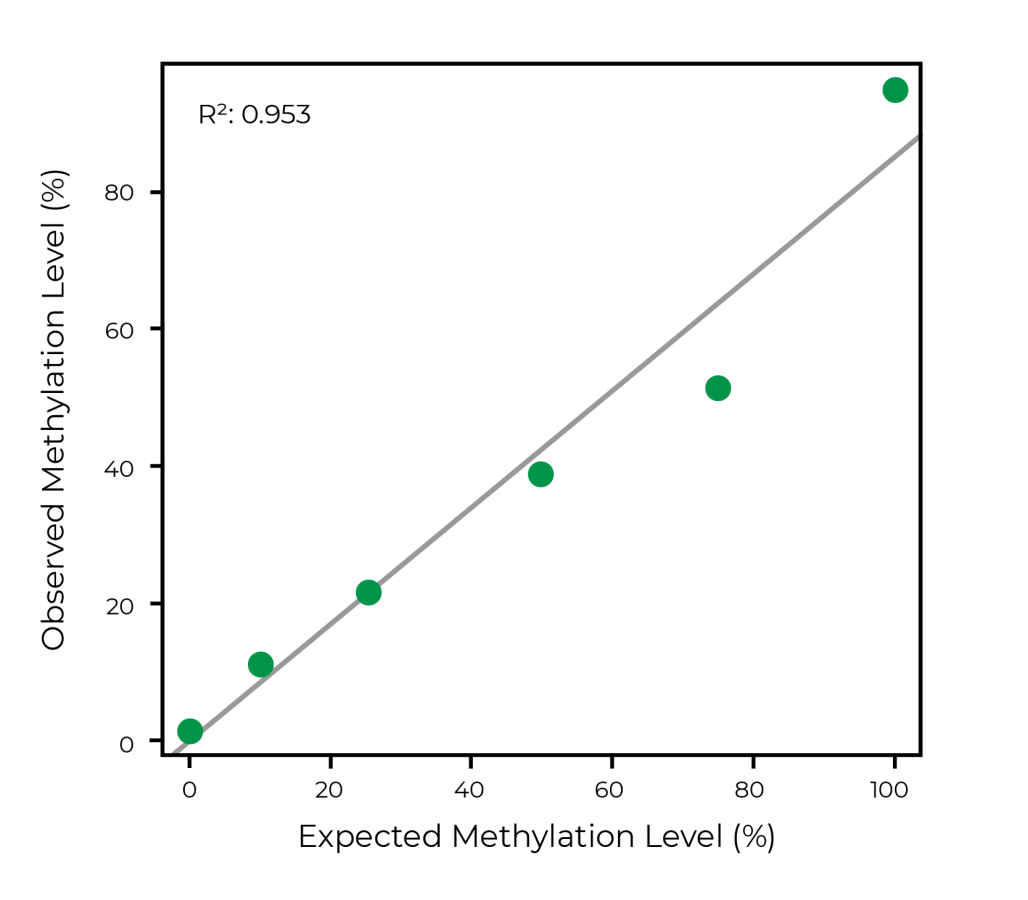
The spike-in control is composed of 6 unique double-stranded synthetic amplicons that have specific DNA methylation levels ranging from 0 to 100%. The observed methylation levels of this control is highly correlated to the expected level when using WGBS. Analysis of this control alongside every sequencing run ensure high quality data for every sample.
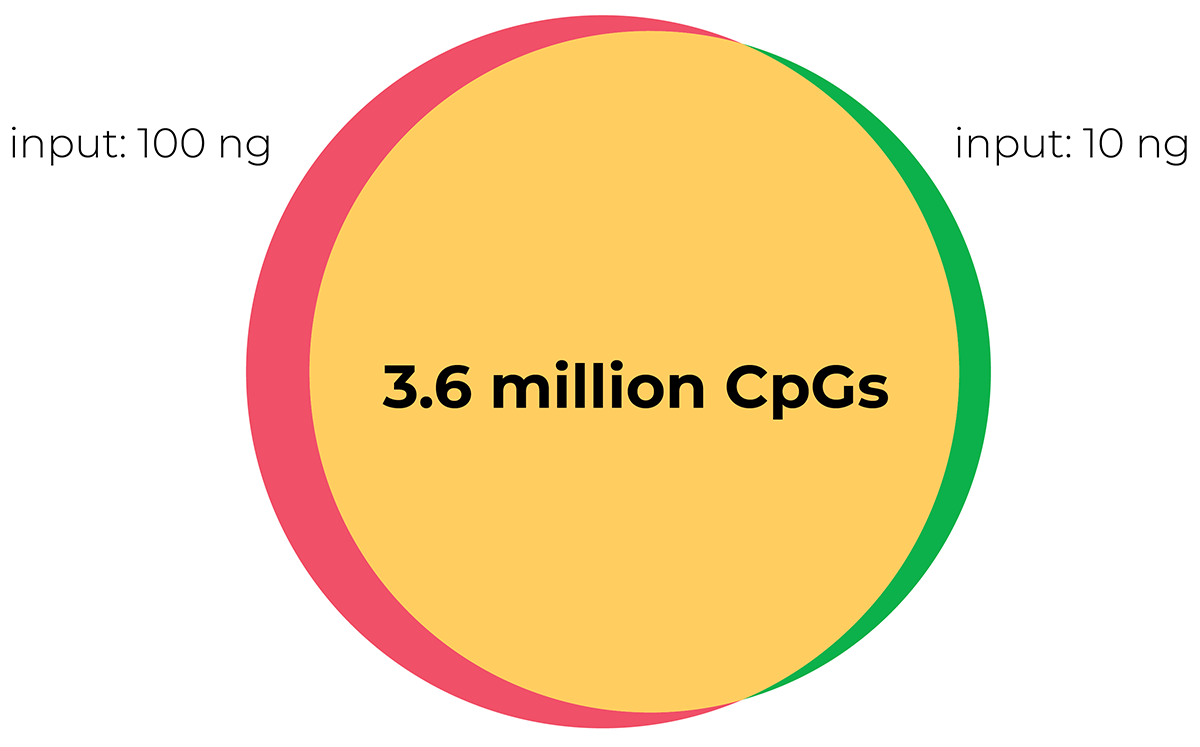
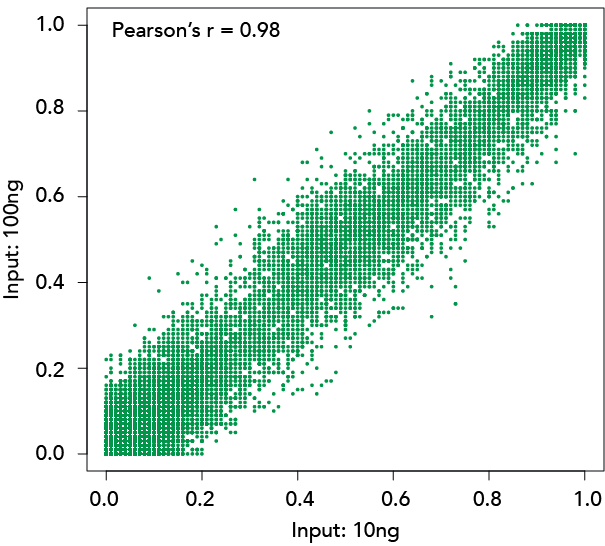
Compatible with low input samples
|
|
|
|---|
Inputs of 10ng and 100ng were subjected to the RRBS workflow and it was shown that the majority of CpG islands analysed in both input types overlapped with each other, and that the methylation ratio was highly correlated between the two.