The LUTHOR High-Definition Single-Cell 3′ mRNA-Seq Library Prep Kit enables high-definition scRNA-seq thanks to Lexogen's Thor amplification technology. With this technology, even low-copy genes are captured and sequenced, leading to a full picture of a cell's gene expression landscape. This facilitates the characterisation of cell sub-populations and makes it the perfect tool for amplifying low-input amounts of RNA material. The kit detects virtually every single mRNA molecule present in the cell to provide a full understanding of each cell's gene expression status, redefining how we approach single-cell RNA sequencing.
Thor technology directly amplifies only the original RNA molecule and produces single fragments per amplified antisense RNA copy. In an efficient one-step library preparation, this technology eliminates the need for lengthy normalisation and allows for straightforward determination of gene expression values at a single cell level.
Benefits of using the LUTHOR HD 3’ mRNA-Seq Library Prep Kit
- Performs exceptionally with input samples as low as 1 pg
- Detect up to 95% of genes at only 1M read depth
- See even lowly expressed genes (< 10 copies/cell) with unmatched sensitivity
- Avoid any read count duplicates with UMI inclusion
- Observe outstanding sequencing statistics with minimal rRNA/gDNA reads
Applications of LUTHOR HD
- Gene expression analysis of purified RNA amounts from 1 ng down to 1pg
- Single-cell differential gene expression analysis
- Sub-population characteristion after 10x genomics experiments
- Rare cell RNA analysis (e.g. circulating tumour cells (CTCs))
- LIVE-Seq (extraction of cytoplasm at different time points)
Detect up to 95% of genes at only 1M read depth, both with isolated single cells and equivalent amounts of purified RNA
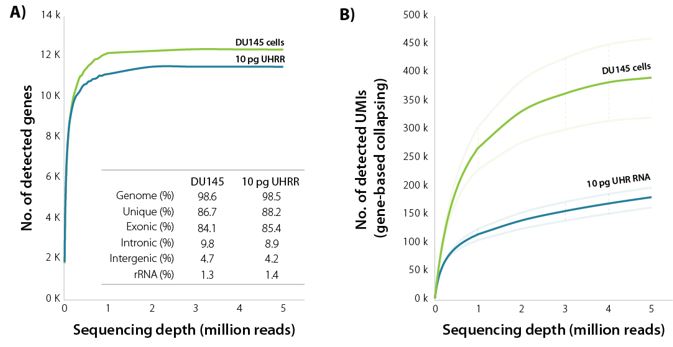
Figure 1 | Sensitivity of LUTHOR HD. A) Scatter plots of the average number of genes detected per DU145 human cell (contains 18.3 ± 1.5 pg of total RNA) and 10 pg Universal Human Reference RNA (UHRR), inferred across four replicates at stepwise-reduced read fractions (CPM > 1). Inset table shows sequencing alignment metrics across four DU145 and four 10 pg UHRR replicates at 1 M read depth. Variability between replicates is negligible (not shown here).
B) Scatter plots of the average number of UMIs (UMI = 12 bp) detected per DU145 human cell and 10 pg UHRR, inferred across four replicates at stepwise-reduced read fractions. UMI counts represent all unique alignments after gene-based collapsing. The number of transcripts directly correlates with cell size (large cells have more transcripts, small cells have fewer transcripts).
Outstanding sequencing statistics- minimal rRNA and gDNA reads
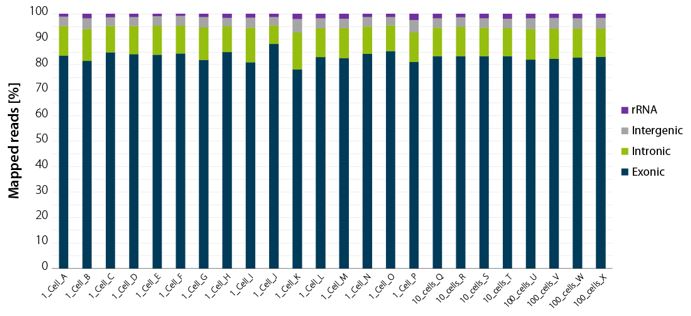
Figure 2 | LUTHOR HD NGS read statistics. Horizontal stacked bar graph shows sequencing alignment metrics across 16 single DU145 cells (named A to P), 4 groups of 10 cells (named Q to T), and 4 groups of 100 cells (named U to X), freshly isolated by flow-cytometry-assisted sorting and immediately lysed in LUTHOR HD Cell Lysis Buffer (CLB). Read depth per library ranges from 0.44 to 1 M reads (1 cell), 0.78 to 1 M reads (10 cells) and 0.80 to 1 M reads (100 cells). Total mapped reads percentages are split into exonic, intronic, intergenic, and ribosomal RNA reads.
LUTHOR HD consistently delivers high reproducibilty, even when repeated by different operators and at ultra-low input
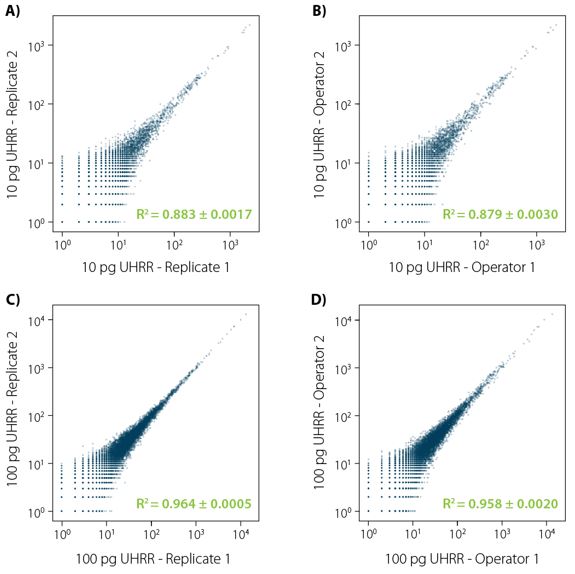
Figure 3 | LUTHOR HD reproducibility. Correlation plots (orthogonal regression of gene-based collapsed and mapped reads) obtained from UHRR reference samples. Four different comparisons were conducted: A) same operator, 4 technical replicates (6 comparisons), 10 pg input; B) two different operators, 4 technical replicates per operator (16 comparisons); C) same as A, with 100 pg input; D) same as C, with 100 pg input. Each image illustrates an exemplary comparison of two samples. Variance (±) indicated for the R² value is the result of all combinations in each comparison experiment.
System compatibility
- HiSeq® 2000/2500/3000/4000
- NextSeq® 500/550/2000
- MiSeq®
- iSeq®
- NovaSeq® 6000
Material available for download
Luthor HD Single-Cell 3’ mRNA-Seq Library Prep kits flyer
Luthor HD Single-Cell 3’ mRNA-Seq Library Prep user guide
HiSeq, NextSeq, MiSeq, iSeq and NovaSeq are registered trademarks of Illumina, Inc.
Products
Note: product availability depends on country - see product detail page.
| Details | Cat number & supplier | Size | Price |
| LUTHOR High-Definition Single-Cell 3' mRNA-Seq Kit 204.24 · Lexogen | 204.24 Lexogen |
24 preparations |
£1748.00
24 preparations
view
|
| LUTHOR High-Definition Single-Cell 3' mRNA-Seq Kit 204.96 · Lexogen | 204.96 Lexogen |
96 preparations |
£6443.00
96 preparations
view
|
| LUTHOR High-Definition Single-Cell 3' mRNA-Seq Kit with UDI 12 nt Set B1, (UDI12B_0001-0024) 221.24 · Lexogen | 221.24 Lexogen |
24 preparations |
£1768.00
24 preparations
view
|
| LUTHOR High-Definition Single-Cell 3' mRNA-Seq Kit with UDI 12 nt Set B1, (UDI12B_0001-0096) 221.96 · Lexogen | 221.96 Lexogen |
96 preparations |
£6542.00
96 preparations
view
|